Regenerative random forest with automatic feature selection
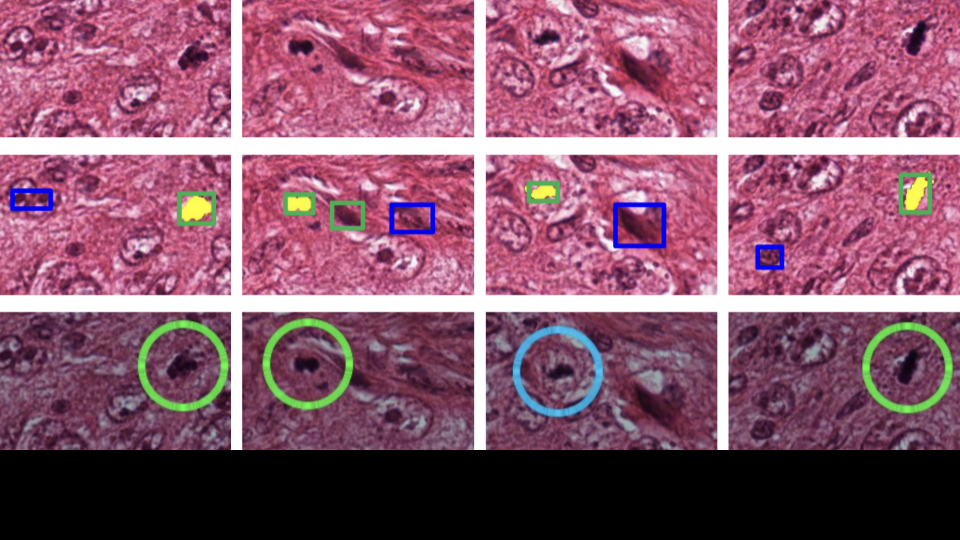
Detecting and counting of mitotic cells from histopathological images are key steps in breast cancer diagnosis as mitosis signals cell division. Interest in automating this process is driven from the arduous and error-prone nature of the manual detection. Research in this area has received much interest after the launch of mitosis detection as a challenge in ICPR 2012. The images that need to be analyzed are obtained with Hematoxylin & Eosin (H&E) staining which render the cell nuclei dark blue against pinkish background.
Early solutions proposed for this problem include Gamma-Gaussian mixture modeling and independent component analysis. The difficulty in identifying appropriate features has been addressed by augmenting handcrafted features with those learnt by a convolutional neural network or using only feature learnt with a deep neural network. But this benefit of neural networks comes at a cost of tuning effort and training time for optimal performance. For instance, a study reports a training time of 1 day on a GPU and processing time of 8 minutes per image. The latter poses a major deterrent for considering automated mitosis detection with high throughput processing of tissue microarrays. More extensive review can be found.
Recently, random forests with population update have been tried for mitosis detection where tree weights are changed based on classification performance. An in-depth biological study of the breast cancer tissues reveal that key factors in mitosis detection are: color of the nucleus, shape of the nuclear membrane and texture of the surrounding region. Presence of nucleus in a stromal region rules out the possibility of it being mitotic, while the absence or rupture of the nuclear membrane signals mitosis.
We propose a fast mitosis detection method with the following contributions (i) new features based on domain knowledge mentioned above and (ii) regenerative random forest-based classification with automatic feature selection, unlike earlier works. Automatic feature selection is achieved using a novel feature weighting scheme. Feature weights are based on the importance of a feature and we reject features with low weights. A new generation of forest (new population of trees) is created which operates on a reduced feature set. During the test phase, each tree of the trained forest votes with its corresponding weights to perform the classification. We prove that the regeneration process converges producing maximum classification accuracy during training. We achieve lower time complexity comp ared to state-of-the-art techniques.