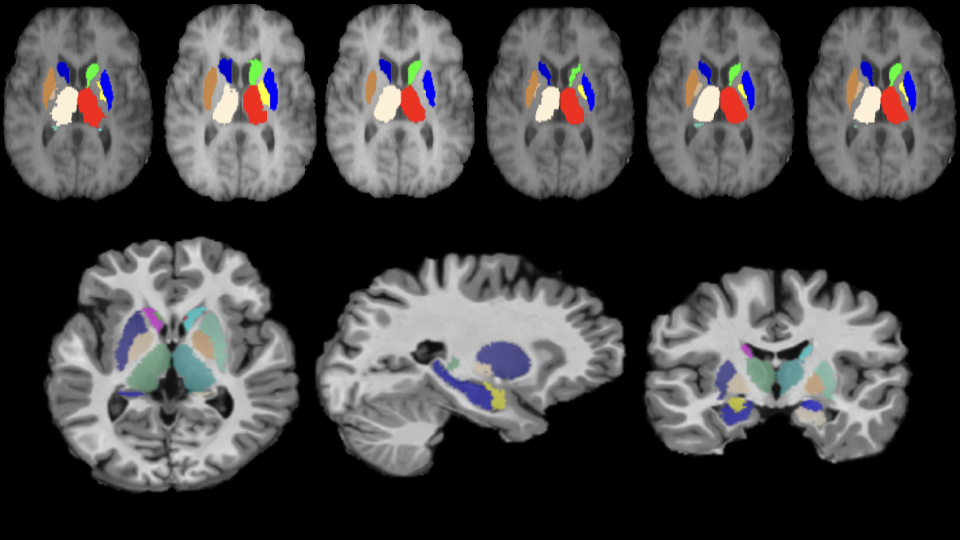
A convolutional neural network for deep brain structures segmentation
Diagnosis of neuro-degenerative diseases, analysis of development of neonatal brain etc., rely heavily on quantitative and qualitative measurements of different human brain structures, especially deep brain structures. For instance, morphometry and volumetry of hippocampus plays an important role in Alzheimer’s disease assessment. Thus, segmentation of these deep brain structures, in less time, is critical. This problem is generally solved either using Non-rigid Registration or Model based techniques, with both relying on training atlases (with manual segmentation) to segment new volumes, albeit in different ways. The former class of techniques label a new volume by registering (non-rigid) training atlases to it and then fusing the propagated labels whereas, the latter techniques predict the labels from a mathematical model learnt from the training atlases. Model based techniques typically require 15-20 minutes to label new volumes as again.
In this paper, we propose an end-to-end trainable Convolutional Neural Network (CNN) architecture called the M-net, for segmenting deep (human) brain structures from Magnetic Resonance Images (MRI). A novel scheme is used to learn to combine and represent 3D context information of a given slice in a 2D slice. Consequently, the M-net utilizes only 2D convolution though it operates on 3D data, which makes M-net memory efficient. The segmentation method is evaluated on two publicly available datasets and is compared against publicly available model based segmentation algorithms as well as other classification based algorithms such as Random Forrest and 2D CNN based approaches. Experiment results show that the M-net outperforms all these methods in terms of dice coefficient and is at least 3 times faster than other methods in segmenting a new volume which is attractive for clinical use.
A method to remove size bias in sub-cortical structure segmentation
Segmentation and analysis of sub-cortical structures is of interest in diagnosing some neurological diseases. Segmentation is a challenging task because of brain tissue ambiguity and data scarcity. Deep learning (DL) solutions are widely used for this purpose by considering the problem as a semantic segmentation of the brain. In general, DL approaches exhibit a bias towards larger structures when training is done on the whole brain.
We propose a method to address this problem wherein a pre-training step is used to learn tissue characteristics and a rough ROI extraction step aids focusing on local context. We use a Residual U-net for demonstrating the proposed method. Experiments on the IBSR and MICCAI datasets show that our proposed solution leads to an improvement in segmentation performance in general with medium and small size structures benefiting significantly. The performance with the proposed method is also marginally better than a more complex, state of art sub-cortical structure segmentation method. A strength of the proposed method is that it can also be applied as a modification to any existing segmentation solution.