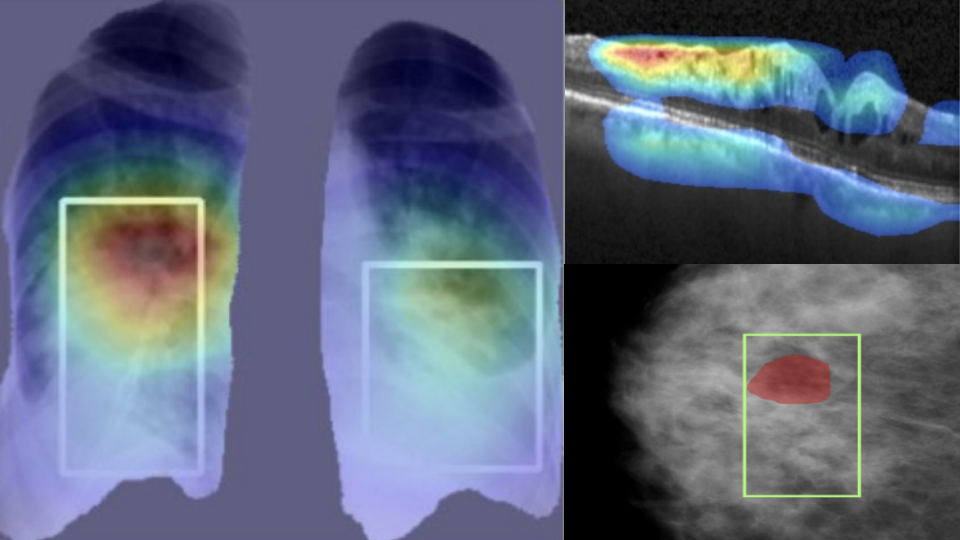
COVID detection from chest X-Ray images with clinically consistent explanations
Deep learning based methods have shown great promise in achieving accurate automatic detection of Coronavirus Disease (covid-19) from Chest X-Ray (cxr) images. However, incorporating explainability in these solutions remains relatively less explored. We present a hierarchical classification approach for separating normal, non- covid pneumonia (ncp) and covid cases using cxr images. We demonstrate that the proposed method achieves clinically consistent explainations. We achieve this using a novel multi-scale attention architecture called Multi-scale Attention Residual Learning (marl) and a new loss function based on conicity for training the proposed architecture.
The proposed classification strategy has two stages. The first stage uses a model derived from DenseNet to separate pneumonia cases from normal cases while the second stage uses the marl architecture to discriminate between covid and ncp cases. With a five-fold cross validation the proposed method achieves 93%, 96.28%, and 84.51% accuracy respectively over three large, public datasets for normal vs. ncp vs. covid classification. This is competitive to the state-of-the-art methods. We also provide explanations in the form of GradCAM attributions, which are well aligned with expert annotations. The attributions are also seen to clearly indicate that marl deems the peripheral regions of the lungs to be more important in the case of covid cases while central regions are seen as more important in ncp cases. This observation matches the criteria described by radiologists in clinical literature, thereby attesting to the utility of the derived explanations.
Lung nodule malignancy classification with weakly supervised explanation
Explainable AI aims to build systems that not only give high performance but also are able to provide insights that drive the decision making. However, deriving this explanation is often dependent on fully annotated (class label and local annotation) data, which are not readily available in the medical domain. This paper addresses the above-mentioned aspects and presents an innovative approach to classifying a lung nodule in a CT volume as malignant or benign, and generating a morphologically meaningful explanation for the decision in the form of attributes such as nodule margin, sphericity, and spiculation. A deep learning architecture that is trained using a multi-phase training regime is proposed. The nodule class label (benign/malignant) is learned with full supervision and is guided by semantic attributes that are learned in a weakly supervised manner.
Results of an extensive evaluation of the proposed system on the LIDC-IDRI dataset show good performance compared with state-of-the-art, fully supervised methods. The proposed model is able to label nodules (after full supervision) with an accuracy of 89.1% and an area under curve of 0.91 and to provide eight attributes scores as an explanation, which is learned from a much smaller training set. The proposed system’s potential to be integrated with a sub-optimal nodule detection system was also tested, and our system handled 95% of false positive or random regions in the input well by labeling them as benign, which underscores its robustness. The proposed approach offers a way to address computer-aided diagnosis system design under the constraint of sparse availability of fully annotated images.
Explainable disease classification via weakly-supervised segmentation
Deep learning based approaches to Computer Aided Diagnosis (CAD) typically pose the problem as an image classification (Normal or Abnormal) problem. These systems achieve high to very high accuracy in specific disease detection for which they are trained but lack in terms of an explanation for the provided decision/classification result. The activation maps which correspond to decisions do not correlate well with regions of interest for specific diseases. This paper examines this problem and proposes an approach which mimics the clinical practice of looking for an evidence prior to diagnosis.
A CAD model is learnt using a mixed set of information: class labels for the entire training set of images plus a rough localisation of suspect regions as an extra input for a smaller subset of training images for guiding the learning. The proposed approach is illustrated with detection of diabetic macular edema (DME) from OCT slices. Results of testing on a large public dataset show that with just a third of images with roughly segmented fluid filled regions, the classification accuracy is on par with state of the art methods while providing a good explanation in the form of anatomically accurate heatmap /region of interest. The proposed solution is then adapted to Breast Cancer detection from mammographic images. Good evaluation results on public datasets underscores the generalisability of the proposed solution.