Efficient and Generic Interactive Segmentation Framework to Correct Mispredictions during Clinical Evaluation of Medical Images
IIIT Hyderabad IIT Delhi
MICCAI, 2021
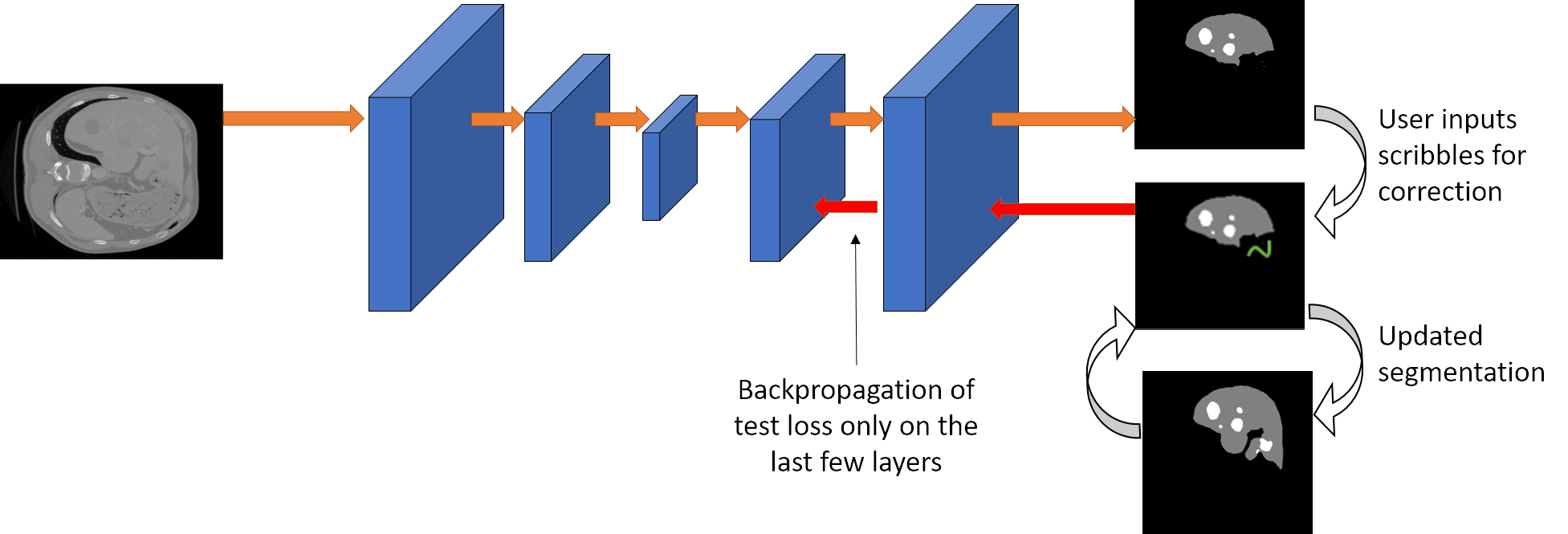
We propose a novel approach to generate annotations for medical images of several modalities in a semi-automated manner. In contrast to the existing methods, our method can be implemented using any semantic segmentation method for medical images, allows correction of multiple labels at the same time and addition of missing labels.
Abstract
Semantic segmentation of medical images is an essential first step in computer-aided diagnosis systems for many applications. However, modern deep neural networks (DNNs) have generally shown inconsistent performance for clinical use. This has led researchers to propose interactive image segmentation techniques where the output of a DNN can be interactively corrected by a medical expert to the desired accuracy. However, these techniques often need separate training data with the associated human interactions, and do not generalize to various diseases, and types of medical images. In this paper, we suggest a novel conditional inference technique for deep neural networks which takes the intervention by a medical expert as test time constraints and performs inference conditioned upon these constraints. Our technique is generic can be used for medical images from any modality. Unlike other methods, our approach can correct multiple structures at the same time and add structures missed at initial segmentation. We report an improvement of 13.3, 12.5, 17.8, 10.2, and 12.4 times in terms of user annotation time compared to full human annotation for the nucleus, multiple cell, liver and tumor, organ, and brain segmentation respectively. In comparison to other interactive segmentation techniques, we report a time saving of 2.8 , 3.0, 1.9, 4.4, and 8.6 fold. Our method can be useful to clinicians for diagnosis and, post-surgical follow-up with minimal intervention from the medical expert.
Paper
Additional Details
Some additional details have been provided which we were unable to put in the paper due to space constraints.
Qualitative Results
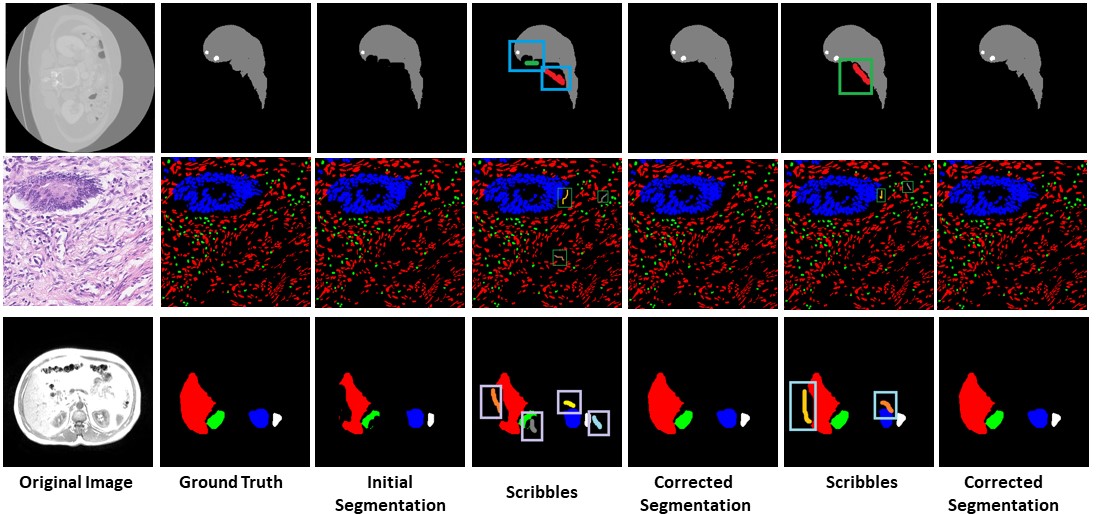
Multiple Label Segmentation
Our approach has the capability to interactively correct the segmentation of multiple labels at the same time
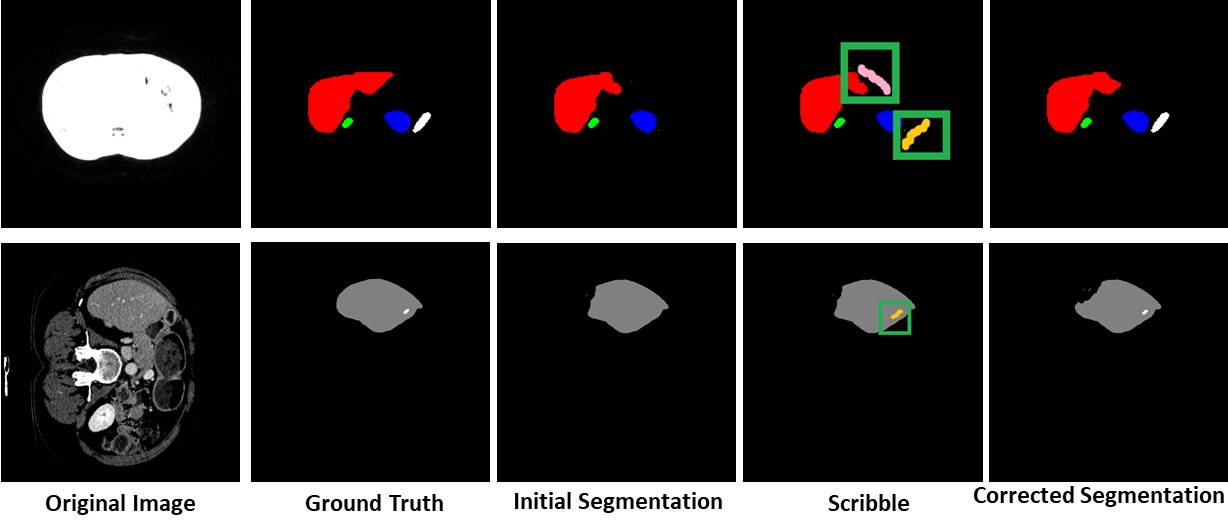
Missing Label Segmentation
Our method has the capability to add labels missed at the initial segmentation.
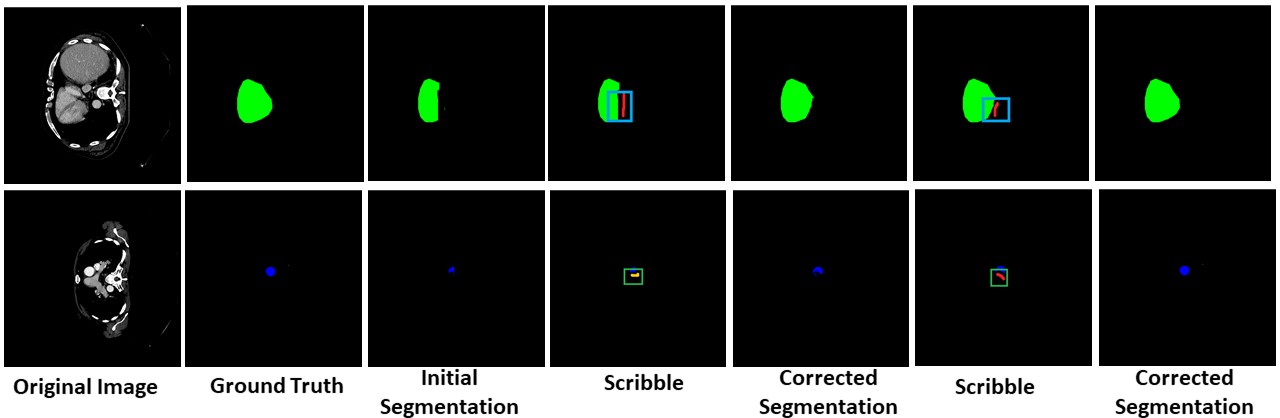
Unseen Organ Segmentation
We can perform interactive segmentation of organs for which the pre-trained model was not trained for.
Network Details
The details of the networks used in our paper has been given here.
The network is based on the DRIU architecture. It is a cascaded architecture where the liver is segmented first followed by the lesion.
A multiple branch network has been proposed which does nuclear instance segmentation and classification at the same time. The horizontal and vertical distances of nuclear pixels between their centers of masses are leveraged to separate the clustered cells.
An autofocus layer for semantic segmentation has been proposed here. The autofocus layer is used to change the size of the receptive fields which is used to obtain features at various scales. The convolutional layers are paralellized with an attention mechanism.
Contact
- Bhavani Sambaturu - This email address is being protected from spambots. You need JavaScript enabled to view it.
- Ashutosh Gupta - This email address is being protected from spambots. You need JavaScript enabled to view it.