A Rule-based Approach to Image Retrieval
Published in: IEEE Region 10 Conference on Convergent Technologies, TENCON 2003
Authors: Dhaval Mehta, E.S.V.N.L.S.Diwakar,, C. V. Jawahar
Published in: IEEE Region 10 Conference on Convergent Technologies, TENCON 2003
Authors: Dhaval Mehta, E.S.V.N.L.S.Diwakar,, C. V. Jawahar
Published in: International Conference on Pattern Recognition, ICPR 2006
Authors: P Suman Karthik, C. V. Jawahar
Retrieval and indexing in multimedia databases has been an active topic both in the Information Retrieval and com- puter vision communities for a long time. In this paper we propose a novel region based indexing and retrieval scheme for images. First we present our virtual textual description using which, images are converted to text documents con- taining keywords. Then we look at how these documents can be indexed and retrieved using modified elastic bucket tries and show that our approach is one order better than stan- dard spatial indexing approaches. We also show various operations required for dealing with complex features like relevance feedback. Finally we analyze the method compar- atively and and validate our approach.
 |
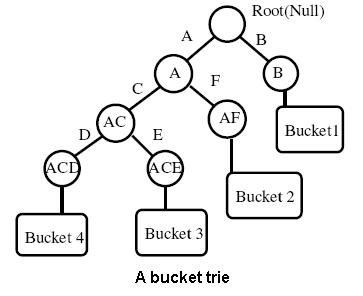 |
Published in: 3rd International Conference on Visual Information Engineering, VIE 2006
Authors: P Suman Karthik, C. V. Jawahar
Abstract:
The state of the art in contemporary visual object categorization and classification is dominated by “Bag Of Words” approaches. These use either discriminative or generative learning models to learn the object or scene model. In this paper, we propose a novel “Bag of words” approach for content based image retrieval. Images are converted to virtual text documents and a new relevance feedback algorithm is applied on these documents. We explain how our approach is fundamentally different to existing ones and why it is ideally suited for CBIR. We also propose a new hybrid relevance feedback learning model. This merges the best of generative and discriminative approaches to achieve a robust and discriminative visual words based description of a visual concept. Our learning model and “Bag Of Words” approach achieve a balance between good classification and efficient image retrieval.
 |
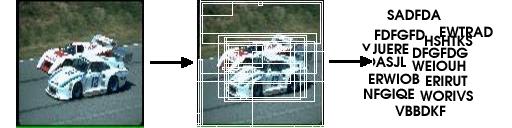 |
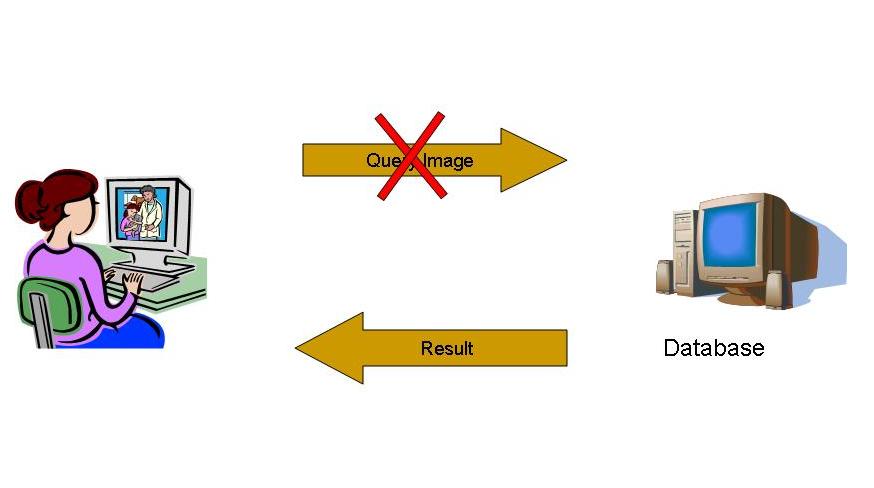
For content level access, very often database needs the query as a sample image. However, the image may contain private information and hence the user does not wish to reveal the image to the database. Private Content Based Image Retrieval (PCBIR) deals with retrieving similar images from an image database without revealing the content of the query image . not even to the database server.
Preprocessing Step
The algorithm is based on the Quadratic Residuosity Assumption which states that:
Given a number `y` belongs to YN, it is predictably hard to decide whether `y` is a QR or a QNR..
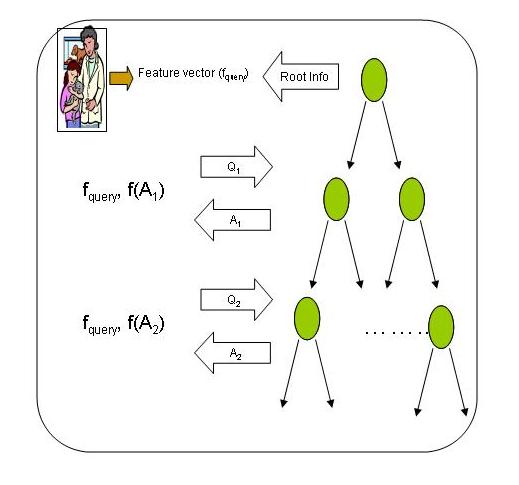
Hierarchical structures primarily vary in
If we can take care of these two, the algorithm can be extended to any hierarchical structure used for indexing. Our algorithm requires a m x n matrix to be formed and a set of nodes at a level can be easily converted in to such a format (irrespective of the number of the nodes). Moreover the data transfer takes place in binary format and any data in the intermediate nodes of an indexing structure can be represented in binary format. So if the user has the data about the indexing structure and the format of the information stored at a node, the algorithm can be simulated for any hierarchical structure.
Experiments were conducted on popular indexing schemesto check the feasibility and applicability of the PCBIR algorithms. The experiments were conducted using a GNU C compiler on a 2 GHz Intel Pentium M process with 4 GigaBytes of main memory. Information regarding the indexing structures and the datasets.
The Corel dataset consisted of 9907 images, scaled to 256 x 128 resolution. The color feature was used to index the images. The feature vector (i.e color histogram) was 768 in length (256 values each for RGB). A KD tree usually stores the ssplit value and split dimension at non leaf nodes. Each value was represented in 32 bits, thus the total information at each intermediate/non-leaf node was 64 bits. Sample results have been shown below. The image with a black box is the query image and the other images are the top 4 retrieved images. The Corel dataset consisted of 9907 images, scaled to 256 x 128 resolution. The color feature was used to index the images. The feature vector (i.e color histogram) was 768 in length (256 values each for RGB). A KD tree usually stores the ssplit value and split dimension at non leaf nodes. Each value was represented in 32 bits, thus the total information at each intermediate/non-leaf node was 64 bits. Sample results have been shown below. The image with a black box is the query image and the other images are the top 4 retrieved images.


The average retrieval time for a query was 0.596 secs. The time was obtained by amortization over 100 queries.
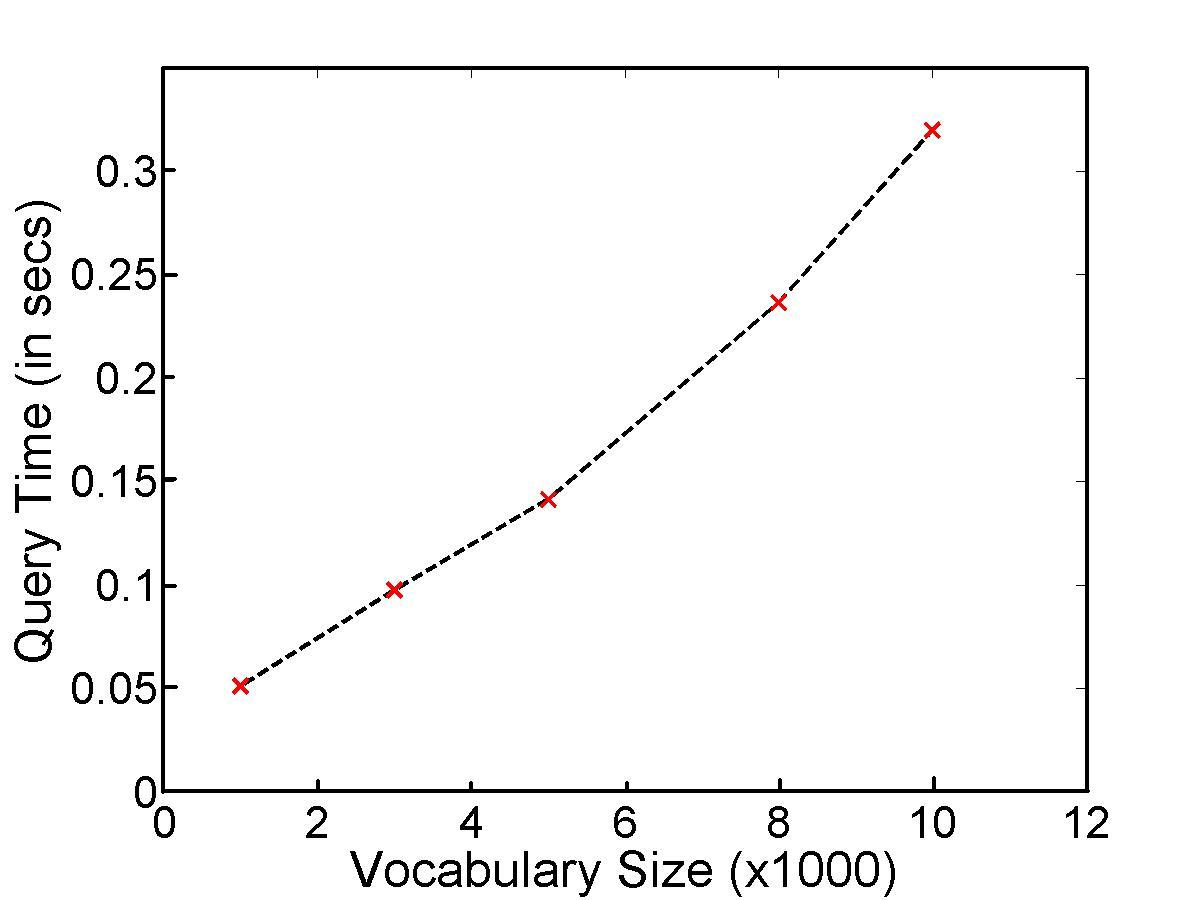
SIFT featues in an image were used to obtain a vocabulary of visual words. The vocabulary was of 10,000 visual words. The Nister dataset consisting of 10,200 images was used. The dataset consists of 2540 object photographed from 4 different angles and lighting conditions. The vocabulary tree was constructed using a training set of 2000 images which were picked from the dataset itself. Sample results have been shown below. The image with a black box is the query image and the other images are the top 4 retrieved images.


The average retrieval time for a query was 0.320 secs. The time was obtained by averaging over 50 queries which were amortized.Since the vocabulary tree allows us to change the size of the vocabulary, retrieval times under various vocabulary sizes were recorded in the following graph.
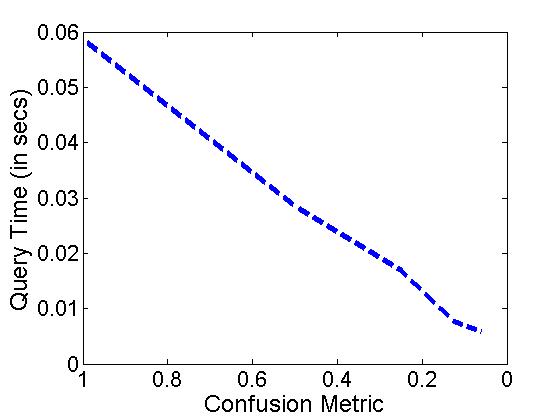
LSH consisted of a set of 90 hash functions each with 450 bins on average. The images in each bin were further subdivided by applying LSH again - Thus gaining an hierarchy of 2 levels. The average retrieval time was 0.221.The algorithm can also be used to achieve partial privacy which is indicated by 'n', also known as the confusion metric. When its value is 1, the retrieval is completely private and 0 indicates that its non-private image retrieval.
 |
 |
Due to the lack of very large standard image datasets, we had to test the scalability of our system using synthetic datasets. The retrieval times for various dataset sizes is provided in the table below.
| Dataset Size | Average Retrieval Time |
| 2^10 | 0.005832 |
| 2^12 | 0.008856 |
| 2^14 | 0.012004 |
| 2^16 | 0.037602 |
| 2^18 | 0.129509 |
| 2^20 | 0.261255 |
Shashank Jagarlamudi, Kowshik Palivela, Kannan Srinathan, C. V. Jawahar : Private Content Based Image Retrieval. IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2008.
| |
Automated management of the content rich and voluminous multimedia data has attracted tremendous attention over the last decades. Any promising solution should ideally have easy web based access with intuitive querying methods and should be designed to respond with majority of relevant results within a few fractions of a seconds, from amongst millions of real images. This requires the building blocks of the solution like the user interface, representation and indexing, comparison and retrieval, learning and memorization, result presentation etc. all to perform together. Literature abounds with work focusing on small subsets of these blocks but a work unifying the blocks is missing. A practical solution requisites optimal integration all the blocks.
In FISH we present a complete end-to-end practical system for fast image search in huge databases FISH uses a query-by-example based simplistic interface which maximizes user input. Our proposal uses a set of standard MPEG-7 visual descriptors for representing images. We achieve this with a highly adaptive B+ - tree based indexing scheme which supports the inherent organization of data into similarity clusters. We also adopt a set of popular relevance feedback approaches for incremental short term learning and propose a highly suited scheme for inexpensive and adaptive inter-query or long term learning. We also use the long term learning based long term memory for adaptive improving ROI extraction from images. In our experiments FISH demonstrated commendable retrieval performance from a million real images within fractions of a second.
We present here the results from some of our major experiments while redirect the reader to the paper for a detailed discussion.
 |
 |
| Learning based improvement across feedback iterations over a randomly picked train query | |
 |
 |
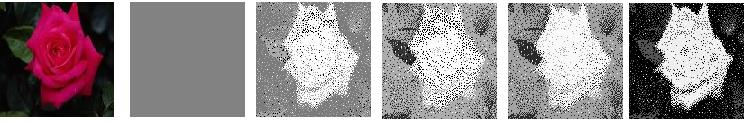
| Long term memory based extraction of ROI shown for a few sample images from the database. | |
 |
|
| Paper [pdf] | Demo |